Simoa Dry Blood Extraction Kit
/in Partners, Quanterix/by Harshita Sharma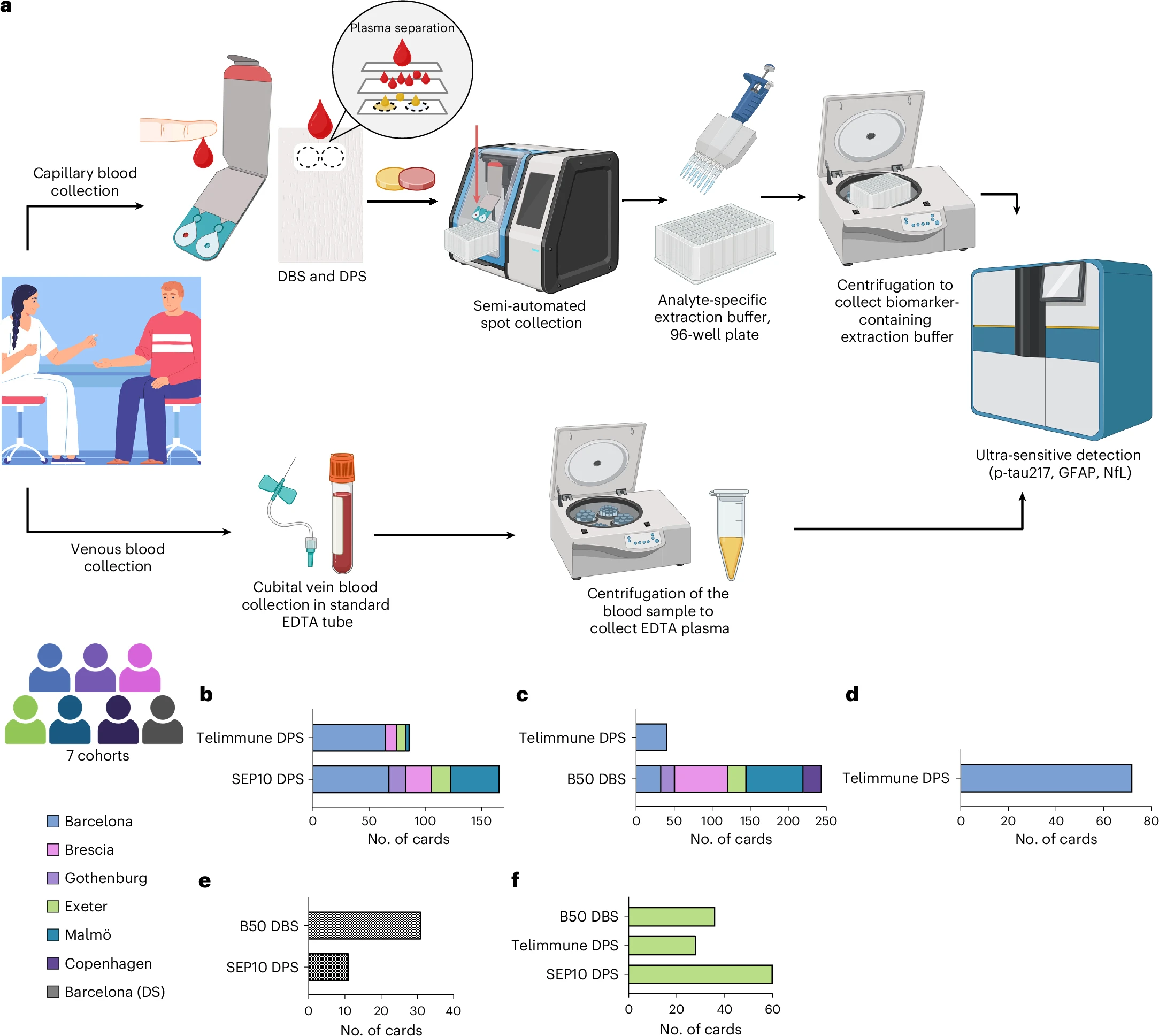
Huber, H., Montoliu-Gaya, L., Brum, W.S. et al. A minimally invasive dried blood spot biomarker test for the detection of Alzheimer’s disease pathology. Nat Med (2026). https://doi.org/10.1038/s41591-025-04080-0
The Simoa Dry Blood Extraction Kit is a validated, device-agnostic solution for extracting analytes from dried plasma and dried blood spot (DPS/DBS) samples for use with Simoa® assays. It enables consistent, reproducible recovery of low-abundance biomarkers while maintaining the femtogram-level sensitivity required for clinical and translational research.
Designed to support decentralized and remote sample collection, the kit simplifies pre-analytical workflows without compromising data quality. It is well suited for longitudinal studies, multi-site trials, and settings where traditional venous sampling and cold-chain logistics are impractical.
Device-agnostic compatibility
Validated across multiple dried plasma and dried blood spot collection devices, enabling consistent analyte recovery independent of collection format. This supports reliable data generation across decentralized, remote, and longitudinal study designs.
Standardized, semi-automated workflow
A harmonized extraction protocol controls critical pre-analytical variables, including elution conditions, centrifugation parameters, and buffer composition. This reduces operator- and site-dependent variability and improves reproducibility across batches and study sites.
Preservation of analytical sensitivity
The extraction process is optimized to maintain the ultra-high sensitivity of Simoa® assays, enabling reliable detection of low-abundance biomarkers such as p-Tau 217. Suitable for neurological, inflammatory, and systemic biomarker applications where signal integrity is critical.
Direct compatibility with Simoa® assays
Designed specifically for downstream use with Simoa® kits, the workflow integrates into existing laboratory processes without the need for additional assay optimization or protocol development.

Hanna Huber
Nutrition Scientist // Ph.D // Postdoctoral researcher @DZNE Bonn & University of Gothenburg
The Simoa Dry Blood Extraction Kit workflow is supported by evidence from the DROP-AD study, published in Nature Medicine, demonstrating that capillary dried blood samples can be used to reliably quantify key Alzheimer’s disease biomarkers.
The multi-centre European study showed strong concordance between capillary dried blood and conventional venous plasma measurements for:
-
p-Tau217 – strong correlation across multiple sites
-
GFAP and NfL – reliable quantification with high plasma concordance
The approach demonstrated good diagnostic accuracy for identifying CSF-confirmed Alzheimer’s pathology and high reproducibility with self-collected samples, supporting decentralized and remote study designs.
Importantly, the study confirms the feasibility of this approach in high-risk and underrepresented populations, including individuals with Down syndrome, and eliminates the need for venipuncture, centrifugation, and cold-chain logistics.
This workflow is intended for research use only and is not designed for clinical diagnosis or clinical decision-making.
Unlock with quick sign up!
The Simoa Dry Blood Extraction Kit is designed to support high-sensitivity biomarker analysis in settings where traditional venous sampling and cold-chain logistics are limiting.
For academic and translational neuroscience teams conducting large cohort or longitudinal studies, the kit provides a standardized method for extracting biomarkers from small-volume, remotely collected samples, reducing pre-analytical variability and improving inter-site comparability.
In pharmaceutical and biotechnology research, the workflow supports early-phase and decentralized study designs where sample volume is limited but analytical sensitivity is critical.
For public health and global research settings, the kit enables reliable laboratory-grade biomarker analysis from capillary dried blood samples, supporting studies in low-resource or geographically distributed populations.
The Simoa Dry Blood Extraction Kit is intended for the extraction of analytes from capillary-derived dried blood and plasma collection devices to support detection of low-abundance biomarkers using Simoa® assay kits.
For research use only.
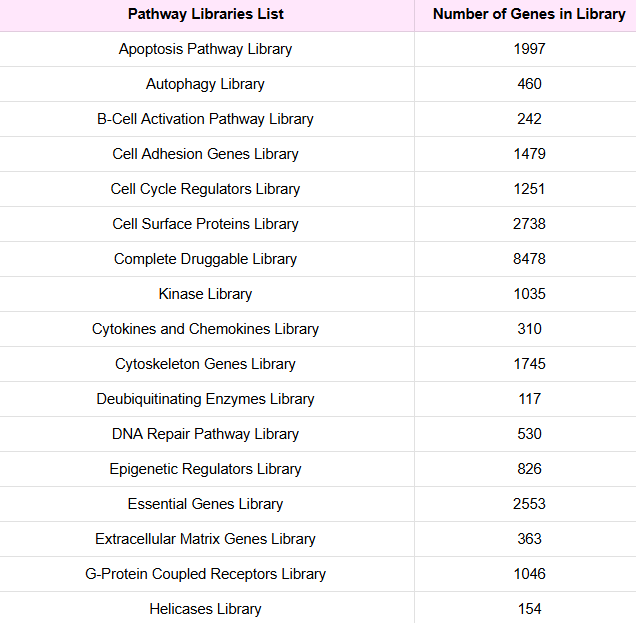
Kit Contents
Each kit includes Quanterix extraction buffer and two precipitation plates.

Julia Young
Quanterix Business Manager
As the official distributor of Quanterix in Australia and New Zealand, Decode Science is providing local access to Simoa® platforms, assays, and workflow solutions with region-based technical and application support.